Stable Isotopes for Mass Spectrometry-based Quantitative Proteomics
Quantitative proteomics is a powerful technique for analyzing protein dynamics in biological systems, including both discovery and targeted approaches. This method primarily uses isotopic labeling of proteins or peptides, which are then distinguished by mass spectrometry.
Overview
- Quantitative Proteomics Mass Spectrometry
- Stable Isotopes in MS-based Quantitative Proteomics
- Quantitative Proteomics Methods Based On Stable Isotope Labeling
- Label-free Quantitative Mass Spectrometry
- Discovery and Targeted Proteomics using Mass Spectrometry
- Stable Isotope Application in Metabolomics
Quantitative Proteomics Mass Spectrometry
The basic principle of quantitative analysis by mass spectrometry is the measurement of a signal representative of the mass of the analyte relative to a known amount of an internal standard. Although more complex than traditional protein identification, quantitative proteomics is essential for understanding global protein expression and modification.
The field of quantitative proteomics continues to evolve, providing researchers with increasingly sophisticated tools to study protein dynamics in cells, tissues, and organisms. At Silantes, we are pleased to offer a growing and comprehensive portfolio of stable isotope labeled internal standards for both relative and absolute quantification by mass spectrometry.
Selected products in this field (complete product range at bottom of page):
Stable Isotopes in MS-based Quantitative Proteomics
To address the need to determine protein abundance in biological samples, proteomic analysis has evolved from protein identification to precise quantification of proteins and peptides. Two main quantitative proteomic approaches are used in this field – relative quantification and absolute quantification. The basic principle of these methods is stable isotope labeling, which compares peak intensities of isotope pairs. These pairs include ions with native isotopes and those with heavy stable isotopes such as 2H, 13C, 15N, or 18O.
Relative Quantification
Relative quantification involves comparing the abundance of proteins between different samples without determining their exact concentrations. This method focuses on the ratio of protein levels between conditions, which can indicate changes in expression due to various biological factors. Techniques such as SILAC, iTRAQ, and TMT are commonly used for relative quantification. Proteins from different samples are labeled with isotopes that differ in mass, allowing for simultaneous analysis and comparison of protein levels.
Selected products in this field (complete product range at bottom of page):
Absolute Quantification
Absolute quantification aims to determine the exact concentration of proteins in a sample. This method provides an accurate measurement of protein abundance, which is essential for understanding biological processes at a quantitative level. Absolute measurement usually involves using synthetic peptides that have been labeled with a specific isotope, like in the AQUA technique.
Selected products in this field (complete product range at bottom of page):
Quantitative Proteomics Methods Based On Stable Isotope Labeling
There are several quantitative proteomics methods based on stable isotope labeling, each with unique characteristics and applications for MS analysis of proteins. These methods use stable isotope labeling to improve the accuracy and reliability of protein quantification, making them valuable tools in proteomics research.
Read more about iTRAQ, TMT, and SILAC
Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)
SILAC involves the incorporation of stable isotopes of amino acids into proteins during cell culture. Cells are grown in media containing heavy isotopes (e.g. 13C, 15N) and then compared to cells grown in normal media. This method is useful for studying protein dynamics, interactions, and post-translational modifications in cell lines and biological systems.
Selected products in this field (complete product range at bottom of page):
Isotope-Coded Affinity Tagging (ICAT)
ICAT is a mass spectrometry-based technique used to compare protein expression levels between two samples. The isobaric mass tag is covalently attached to cysteine residues in proteins, allowing quantification based on the accurate mass difference between light and heavy isotopes. This method is effective for comparing protein levels between two samples, especially in complex mixtures.
Isobaric Tags for Relative and Absolute Quantitation (iTRAQ)
iTRAQ involves the labeling of peptides with isobaric mass tags that release reporter ions during mass spectrometry. Multiple samples can be analyzed simultaneously. This method is suitable for high-throughput studies and allows multiplexing of samples.
Tandem Mass Tags (TMT)
Similar to iTRAQ, TMT quantitative proteomic analysis involves isobaric labeling of peptides, allowing simultaneous quantification of multiple samples in a single experiment. This method is useful for comparative proteomics and can analyze multiple samples in a single run.
Absolute Quantification using Stable Isotope Dilution (AQUA)
AQUA involves spiking samples with known amounts of stable isotope-labeled peptides. The ratio of labeled to unlabeled peptides is used to determine absolute protein concentrations. This quantitative approach provides accurate quantification of specific analysis of complex protein mixtures.
Isotope-Coded Protein Labeling (ICPL)
ICPL is a powerful quantitative proteomics technique that uses stable isotopes to label proteins for comparative analysis. This method enables high-throughput profiling of proteins in complex biological samples, making it particularly valuable in proteomics research.
Label-free Quantitative Mass Spectrometry
Alternatively, label-free quantitative proteomics analysis allows the identification and quantification of proteins without the use of chemical labels. This method relies on direct measurements from mass spectrometry data using two primary techniques, such as intensity-based methods and spectral counting methods. However, label-free methods are more variable than label-based methods and require more replicates to achieve similar statistical robustness. In addition, they may have difficulty detecting low-abundance proteins and are highly dependent on consistent sample preparation and mass spectrometry analysis.
Read more about Label-Free versus Label-Based Methods
Discovery and Targeted Proteomics using Mass Spectrometry
Mass spectrometry-based proteomics employs isotope-labeled compounds and standards to enhance protein quantification in both discovery and targeted approaches. While both discovery and targeted proteomics leverage isotope-labeled compounds to enhance quantification, their application differs. Discovery proteomics often uses these standards for relative quantification across a broad range of proteins, whereas targeted proteomics employs them for precise, absolute quantification of specific proteins.
Read more about Techniques, Applications, and Challenges
Discovery Proteomics
In discovery proteomics, which aims to identify and quantify a broad range of proteins in an unbiased manner, isotope-labeled standards play a crucial role in improving quantification accuracy and reproducibility. One common approach is the use of a labeled proteome reference as a global internal standard. The use of a labeled proteome reference is particularly advantageous for large-scale studies, as it provides a cost-effective alternative to using individual isotope-labeled peptides for each protein of interest.
Relevant product (complete product range at bottom of page):
Targeted Proteomics
In targeted proteomics, which focuses on the precise quantification of a predefined set of proteins or peptides, isotope-labeled standards are essential for achieving high sensitivity and accuracy. This method allows for the creation of standard curves, enabling precise absolute quantification of specific proteins. It is particularly beneficial for quantifying low-abundance proteins with high specificity and reproducibility.
Relevant product (complete product range at bottom of page):
Stable Isotope Application in Metabolomics
Metabolomics involves the systematic identification and quantification of metabolites, providing insight into the metabolic processes and pathways that occur in biological systems. It provides a direct view of the physiological state, influenced by both genetic and environmental factors, and differs from genomics and proteomics.
Metabolomics data are typically generated using analytical technologies such as high-resolution mass spectrometry. These methods allow the simultaneous detection and quantification of thousands of metabolites, enabling detailed metabolic profiling. Stable isotopes allow the tracing of metabolic pathways and the quantification of metabolic fluxes and play therefore a crucial role in metabolomics.
Read more about the Role of Mass Spectrometry in Metabolomics
Metabolite Identification and Quantification
Stable isotope labeling is used to identify metabolites by creating unique isotopic patterns that can be matched against compound annotation databases. Here, isotopically labeled internal standards help identify metabolites and allow absolute quantification.
For quantification, stable isotope-labeled metabolites serve as internal standards to accurately measure metabolite concentrations using mass spectrometry. At the same time, isotope dilution methods allow absolute quantification by comparing the signal intensities of labeled and unlabeled forms.
Metabolic Flux Analysis
Stable isotope tracers such as 13C-glucose or 13C-glutamine are administered to living systems to track labeled metabolites from cellular metabolism. By measuring labeling patterns and fractions of labeled metabolites, metabolic fluxes can be calculated at cellular and organismal levels. Stable isotope-based metabolomics provides network-constraining information for more precise flux estimates through metabolic networks
Pathway Discovery
Stable isotope tracing allows unambiguous tracking of heavy elements through complex metabolic networks to reconstruct metabolic pathways. Isotope-labeled metabolites can be traced across the entire metabolome to uncover new metabolic transformations and pathways.
-
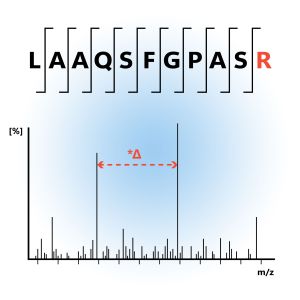
Relative Quantification
-
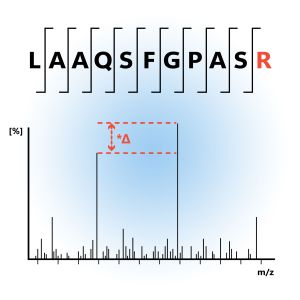
Absolute Quantification
Product Categories:
Products in these categories:
-
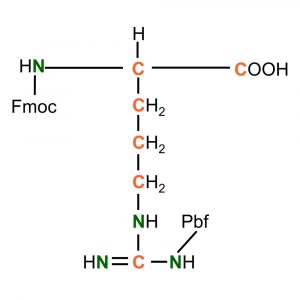
(Fmoc)-L-Arginine(Pbf)-OH
Synonym: L-Arginine, a-N-Fmoc, w-N-Pbf
From: 720 € plus VAT, plus delivery Select options This product has multiple variants. The options may be chosen on the product page
Available in various isotopic labelings and/or quantities. -
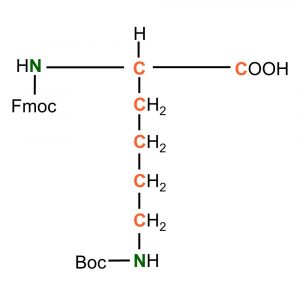
(Fmoc)-L-Lysine(Boc)-OH
Synonym: L-Lysine, a-N-Fmoc, e-N-t-Boc
From: 710 € plus VAT, plus delivery Select options This product has multiple variants. The options may be chosen on the product page
Available in various isotopic labelings and/or quantities. -

2′-O-Methyl(D3)-Adenosine, powder
Synonym: Am-D3
1.090 € plus VAT, plus delivery Add to cart
Quantity: 5mg -

2′-O-Methyl(D3)-Cytidine, powder
Synonym: Cm-D3
1.090 € plus VAT, plus delivery Add to cart
Quantity: 5mg


