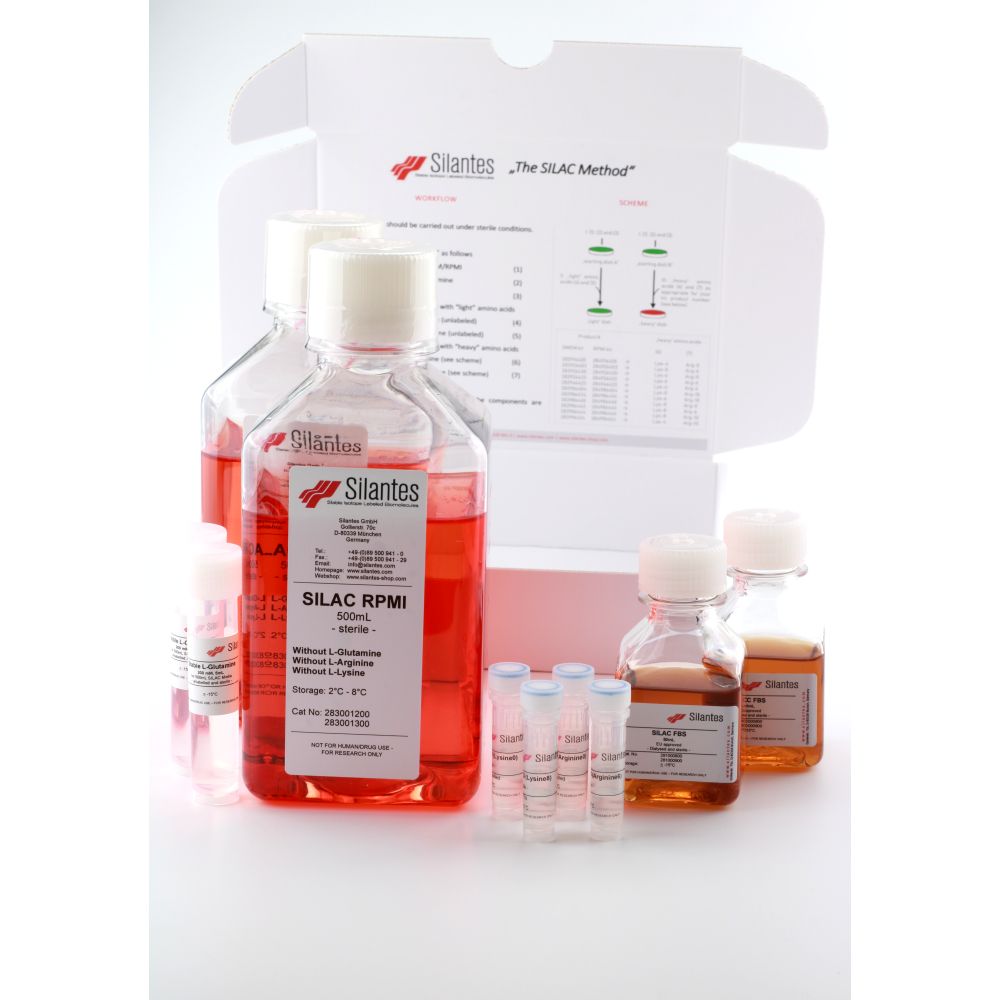
SILAC RPMI Kits
From: 290 € plus VAT, plus delivery
Available in various isotopic labelings.
Quantity: Kit
Description
– Isotopic enrichment for the stable isotope labeled amino acids except for 4, 4, 5, 5, D4 L-lysine: > 98 atom %
– Isotopic enrichment for for 4, 4, 5, 5, D4 L-lysine: > 96 atom %
Dry ice shipment required.
References
| Relevant manuals: | • Silantes Powder RPMI and DMEM for SILAC Preparation of sterile liquid Cell Culture Media: https://silantes.com/documents/manuals/Silantes_SILAC_RPMI_DMEM_Powder_Application_Note.pdf |
|---|---|
| SILAC amino acids from Silantes in scientific publications: | • Hao, B., Li, X., Jia, X., Wang, Y., Zhai, L., Li, D., Liu, J., Zhang, D., Chen, Y., Xu, Y., Lee, S., Xu, G., Chen, X., Dang, Y., Liu, B., & Tan, M. (2020). The novel cereblon modulator CC-885 inhibits mitophagy via selective degradation of BNIP3L. Acta Pharmacologica Sinica, 41(9), 1246–1254. https://doi.org/10.1038/s41401-020-0367-9 • Lößner, C., Warnken, U., Pscherer, A., & Schnölzer, M. (2011). Preventing arginine-to-proline conversion in a cell-line-independent manner during cell cultivation under stable isotope labeling by amino acids in cell culture (SILAC) conditions. Analytical Biochemistry, 412(1), 123–125. https://doi.org/10.1016/j.ab.2011.01.011 • Sigismondo, G., Arseni, L., Palacio-Escat, N., Hofmann, T. G., Seiffert, M., & Krijgsveld, J. (2023c). Multi-layered chromatin proteomics identifies cell vulnerabilities in DNA repair. Nucleic Acids Research, 51(2), 687–711. https://doi.org/10.1093/nar/gkac1264 • Pateetin, P., Hutvagner, G., Bajan, S., Padula, M. P., McGowan, E. M., & Boonyaratanakornkit, V. (2021). Triple SILAC identified progestin-independent and dependent PRA and PRB interacting partners in breast cancer. Scientific Data, 8(1). https://doi.org/10.1038/s41597-021-00884-0 • Lopez-Serra, P., Marcilla, M., Villanueva, A., Ramos-Fernandez, A., Palau, A., Leal, L., Wahi, J. E., Setien-Baranda, F., Szczesna, K., Moutinho, C., Martinez-Cardus, A., Heyn, H., Sandoval, J., Puertas, S., Vidal, A., Sanjuan, X., Martinez-Balibrea, E., Viñals, F., Perales, J. C., . . . Esteller, M. (2014). A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nature Communications, 5(1). https://doi.org/10.1038/ncomms4608 • Ong, S., Kratchmarova, I., & Mann, M. (2002). Properties of 13C-Substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). Journal of Proteome Research, 2(2), 173–181. https://doi.org/10.1021/pr0255708 • Lößner, C., Warnken, U., Pscherer, A., & Schnölzer, M. (2011b). Preventing arginine-to-proline conversion in a cell-line-independent manner during cell cultivation under stable isotope labeling by amino acids in cell culture (SILAC) conditions. Analytical Biochemistry, 412(1), 123–125. https://doi.org/10.1016/j.ab.2011.01.011 •Malet, J. K., Impens, F., Carvalho, F., Hamon, M. A., Cossart, P., & Ribet, D. (2018b). Rapid remodeling of the host epithelial cell proteome by the listeriolysin O (LLO) pore-forming toxin. Molecular & Cellular Proteomics, 17(8), 1627–1636. https://doi.org/10.1074/mcp.ra118.000767 • Rogers, L. C., Kremer, J. C., Brashears, C. B., Lin, Z., Hu, Z., Bastos, A. C., Baker, A., Fettig, N., Zhou, D., Shoghi, K. I., Dehner, C. A., Chrisinger, J. S., Bomalaski, J. S., Garcia, B. A., Oyama, T., White, E. P., & Van Tine, B. A. (2023). Discovery and targeting of a noncanonical mechanism of sarcoma resistance to ADI-PEG20 mediated by the microenvironment. Clinical Cancer Research, 29(16), 3189–3202. https://doi.org/10.1158/1078-0432.ccr-22-2642 • Geiger, T., Wisniewski, J. R., Cox, J., Zanivan, S., Kruger, M., Ishihama, Y., & Mann, M. (2011). Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nature Protocols, 6(2), 147–157. https://doi.org/10.1038/nprot.2010.192 • Hao, B., Sun, M., Zhang, M., Zhao, X., Zhao, L., Li, B., Zhai, L., Liu, P., Hu, H., Xu, J., & Tan, M. (2020). Global characterization of proteome and lysine methylome features in EZH2 wild-type and mutant lymphoma cell lines. Journal of Proteomics, 213, 103614. https://doi.org/10.1016/j.jprot.2019.103614 |
| Relevant blog articles: | • Quantitative Proteomics Explained: Techniques, Applications, and Challenges: https://www.silantes.com/guide-to-quantitative-proteomics-techniques-applications-and-challenges/ • Quantitative Proteomics: Comparing the Big Three – iTRAQ, TMT, and SILAC: https://www.silantes.com/itraq-tmt-silac/ • Quantitative Proteomics: Label-Free versus Label-Based Methods: https://www.silantes.com/label-vs-label-free-quantitative-proteomics/ • Understanding the Role of Mass Spectrometry in Metabolomics: https://www.silantes.com/mass-spectrometry-metabolomics/ • Understanding Proteomics: A Comprehensive Guide to the Various Types: https://www.silantes.com/types-of-proteomics/ |
| Relevant webinars: | • Proteomics using SILAC presented by Prof. Dr. Marcus Krüger: https://www.youtube.com/watch?v=TY6iV8_43iA • Multiplexing with isotopic labels for DDA and DIA in MaxQuant presented by Dr. Jürgen Cox: https://www.youtube.com/watch?v=-Yddv7ehYsk • Stable Isotope Labeling of Mammals presented by Dr. Christoph Turck: https://www.youtube.com/watch?v=BQWvLdILjCA |